Happy Tuesday everyone! Friday was an uneventful day. We started off by cleaning the ITC from our overnight run and sending the data to ourselves so we could work on it in our lab. Cleaning the ITC involves grabbing a liter of Milli-Q Water and putting a needle attached to two hoses in the sample cell. The one hose connects to the liter of Milli-Q and the other to a flask that aspirates the water through the machine. While this was rinsing, we cleaned the injection syringe and the transfer syringe. Our data didn’t have a good baseline, and it was also missing solid peaks. After this, we washed more DNA samples to get them out of the TE buffer and put it in a Sodium Chloride buffer. We did this with 8 samples of 200 microliters of DNA in TE buffer.
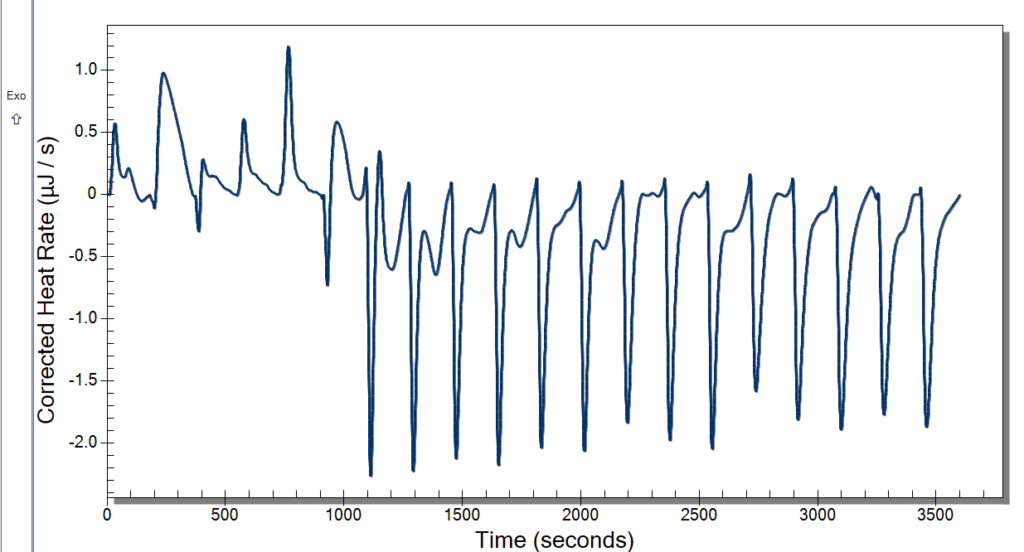
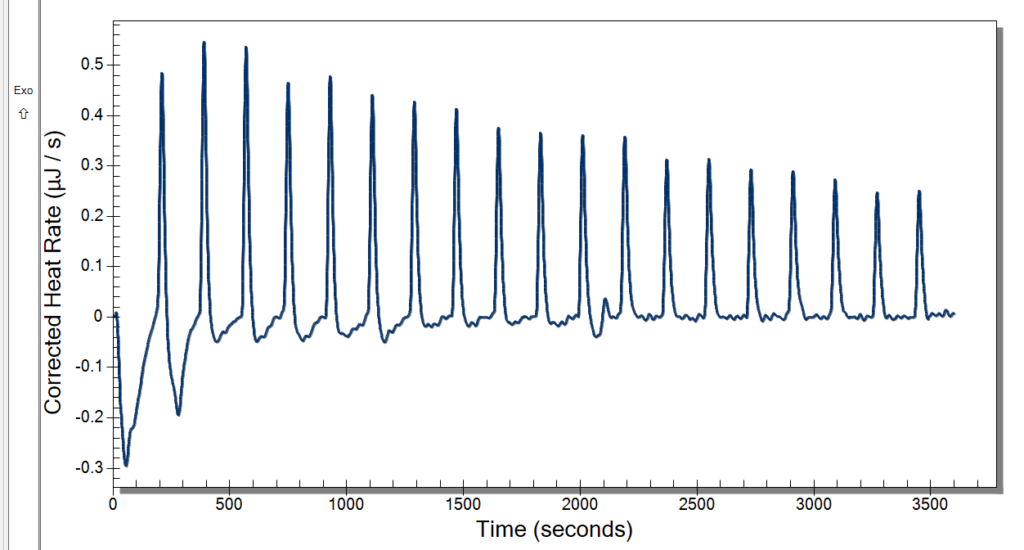
Monday, we had the day off for Memorial Day. Everyone liked having a bit of a break, and I really enjoyed going home and seeing some family. Tuesday, we planned to run the ITC and have our weekly meeting. The time clock also came in, so we had fun messing around with that. It has a very satisfying stamp, so it was a necessary addition to our lab. We started warming the ITC up at 10, so we were able to use it by the time our meeting left out. The meeting involved discussing our progress and plans for the future. We then started our first sample in the ITC then and had 3 mg/mL DNA in 10 mM NaCl for our sample and did 20 injections of 2.5 microliters of our 10 mM Cobalt Hexammine in 10 mM NaCl. Our spin speed was 250 rpm, and the temperature was 25° C. We let this run while we went to our brown bag lunch meetings.

My lunch table had 3 psychology researchers and 2 biology researchers. The psychology researchers couldn’t divulge too much information about their research because they are hoping to use the other X-SIG researchers as participants in their studies, and we could mess up the data by knowing too much about the research beforehand. We are still in the beginning as well, so most of us are just doing the groundwork for our projects and getting trained properly.
After lunch, we returned to some not good-looking data from the ITC. We weren’t certain of what might have caused this, so we cleaned up and did a second run. While that was running, we deduced that it might be too high of a spin rate and that the sample was too small. Originally, we were using the standard spin rate of 250 rpm, but we are going to cut it back to 175 rpm which previous students used. The sample was too small because when it was drawn up into the syringe, air bubbles would enter because we tried to get the last drops from the sample. That is an easy fix though. We will just have to wash more DNA and put it all together.
Macyn also posted our first lab Tik Tok! Go watch it at https://www.tiktok.com/t/ZTRoMh3t7/.



