This week Professor Andresen and I have been preparing mononucleosomes from chicken erythrocytes. Hopefully by Tuesday we will have clean, pure mononucleosomes that we can then use to investigate their electrostatics. So far we have taken the nuclei out of the blood, cleaned it, digested it with micrococcal nuclease and separated the DNA/nucleosomes from the nuclei.
Category: Uncategorized
July 10th – Computer Simulations
Reproducible Results!
| So many dilutions! Endless labelling… |
Last week, I repeated the equilibrium dialysis two separate times and prepared calibrations and dilutions for ICP.
I wasn’t able to run ICP until Monday since we ran out of Argon gas. I ran the ICP on Monday and Tuesday (only 2 runs because my samples were limited). After analyzing the data, we found that both dialysis runs showed 1 Na/S and nearly 1 K/S! I found a calculation error in the data analysis from last week which, when corrected, indicated that both Na and K samples had 1 ion for every 4 S. That dialysis was a “quick and dirty” experiment so we are more confident in the most recent data which has proven to be reproducible.
One thing that looks interesting is that the hydrodynamic radius of the nanoparticles goes down after salt dialysis and back up after water dialysis. After the water dialysis is can be up to 30 nm bigger than after the salt dialysis! Since the amount of PSS/NP is remaining constant, this data indicates that the conformation of the PSS is changing in response to the salt concentration.
| Aggregated Nanoparticles |
Going forward, we are going to run some tests with varying salt concentration. Today I did a titration of NaCl into the PSS coated NPs and took samples at a range of salt concentrations. I found that the nanoparticles aggregated (due to instability) partially at 100mM salt and completely at above 200 mM salt. This will give us an idea of what range we can dialyze the nanoparticles at.
Back the the Lab!
After spending about six months in Australia not doing any physics, I’m back and ready to start a new research project that will hopefully culminate in my capstone project! Instead of working with DNA, I will be conducting experiments using mononucleosomes that Professor Andresen and I will be trying to prepare over the next few days. The protocol looks very scary and full of biochemistry to prepare, clean and inspect the chicken blood until its just nucleosomes but hopefully we will succeed in preparing them ourselves so that it will open many research opportunities in the lab. Wish us luck!
Error Bars
The ICP data from the sodium/potassium trial had extensive error bars due to the large range of sodium and potassium concentrations tested. It was impossible to measure a difference of a few ppm over top of a background concentration of 300 ppm. To resolve this I dialyzed some of the nanoparticles from the previous dialysis with water to clear out the background salt and leave only the gold nanoparticles with anything bound to them. The ICP data for those samples had much smaller error and very interesting results. It showed that there was 1 Na for every 4 sulfurs and 1 K for every 7 sulfurs. Going forward I am going to repeat this process with the water dialysis a few more times get more data and confirm that the data is repeatable.
 |
| Before Water Dialysis |
 |
| After Water Dialysis |
Sodium v Potassium: Round 1
On Monday, I started making a brand new batch of PSS coated NPs. I still have a good portion of the last batch but I wanted to make sure that the data we were seeing was reproducible. On Tuesday, I characterized these particles and ran equilibrium dialysis on them with NaCl and KCl in parallel. This way, I hope to compare the absorption of Na and K ions. Today, I characterized the resultant nanoparticle samples and ran all the samples in the ICP 4 times with 2 different calibrations. If the data is reproducible I hope to see 4 Na per S like last week’s run. The charge on all of the dialyzed nanoparticles since I modified the protocol to dilute with the dialysis buffer has been negative, which matches our original expectations.
June 24th – Data Analysis
For the past week or so I have been working on analyzing the data I gathered from ICP-AES. So far I have found two interesting trends: as the PEG percentage increases (so, DNA-DNA spacing decreases), the charge neutralized by hexamminecobalt(III) increases and the charge neutralized by magnesium decreases, and as the magnesium concentration increases, the charge neutralized by magnesium increases and the charge neutralized by hexamminecobalt(III) decreases. These two trends are not surprising, and correlate well with the expectations of the DNA system. What is even more interesting though, is determining the concentration of magnesium needed at each PEG percentage so that hexamminecobalt(III) and magnesium equally charge neutralize the DNA (shown in the graph below).
ICP
Yesterday, I ran another equilibrium dialysis on the nanoparticles using NaCl. I modified my protocol to dilute the final nanoparticle product with NaCl instead of pure water. Today, I characterized these nanoparticles. They were negatively charged this time which is what we would have expected but we still don’t know why the other two samples are positively charged. This afternoon I am running 4-5 ICP runs on the samples, which I have diluted. This will hopefully give me good statistical data to analyze.
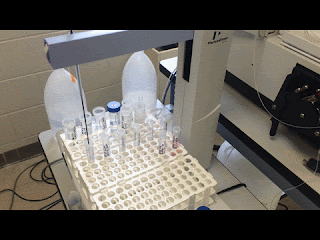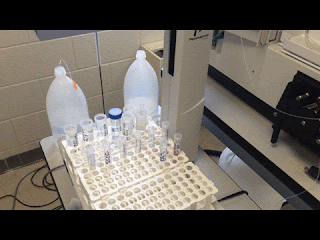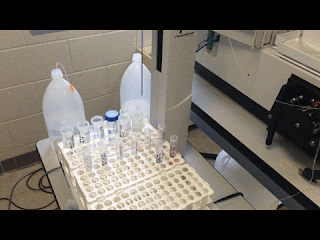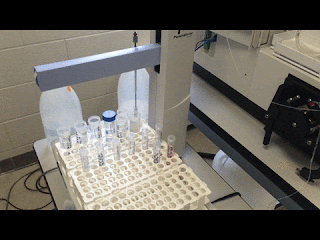 |
| Just watching the autosampler do all my work for me… |
Too much sodium!
Yesterday, I ran another equilibrium dialysis with the same parameters but I doubled the number of spins to 20.
 |
The Files are Breeding Like Tribbles!
On Thursday, I ran ICP on the uncoated nanoparticles and the coated pre-dialysis nanoparticles. Since then, I have spent most of time trying to analyze the ICP data and reading literature on polyelectrolyte deposition. As a result, I have 7 excel documents open on my computer and even more pdfs!
Next week, I plan to run dialysis on the same nanoparticle sample with KCl as the salt buffer. I will then run ICP on those samples so that I can compare them to the nanoparticles that underwent dialysis with NaCl.











