I would like to welcome Diana, Ben, and Nick, three wonderful new students to the lab. We have just begun our adventures together and already they are showing great promise. Keep checking back here for exciting developments as they tell us about their research!
Author: Prof. Andresen
Summer 2014: Magnets, DNA, and Fun!
 |
| Abby and Steve hard at work in our newly created student office! |
The summer has begun with two great new students (and the same old professor). We have a couple of great projects going on this summer from building a new instrument from the ground up to continuing our quest to understand the physics behind DNA packing! (In fact they have gotten some posts in ahead of me below.)
 |
| From Pelta et al., Journal of Biological Chemistry, 1996 |
Abby Bull will be working on the DNA packing part of things. In particular she will be trying to understand the graph at the right. What this is showing is that as cobalthexammine (the chemical is not important, just know that it is a +3 ion) DNA clumps together (aggregates) and falls out of solution. This is surprising because relatively small amounts (~5mM from this graph) of CoHex can do this while there is no amount of +1 or +2 ions that you can add to do the same thing! It is something particular to +3 ions. (Those of you who are observant may also notice that something equally strange happens when you keep adding CoHex.)
Abby will be looking at “pellets” of DNA that are have been put in the state at the bottom of the “valley” in this graph. Ones that have condensed/aggregated/clumped together. Our collaborators at George Washington University have made these samples and then redissolved them in a different solution (in this case a highly concentrated KCl solution). They have made many of these samples, systematically changing the ratio of +3 ion (CoHex) to +1 ion (NaCl). Abby will be measuring the amount of +3 ion and +1 ion that was actually bound to the DNA when it was in its “clumpy” state. Using this, we hope to extract the physics, in particular the forces between DNA, that lead to this state. This is a follow-up on some work done previously by John Giannini in our lab that looked at the amount of +3 ions and +2 ions that were bound in the same state.
 |
| Image courtesy of Wikipedia |
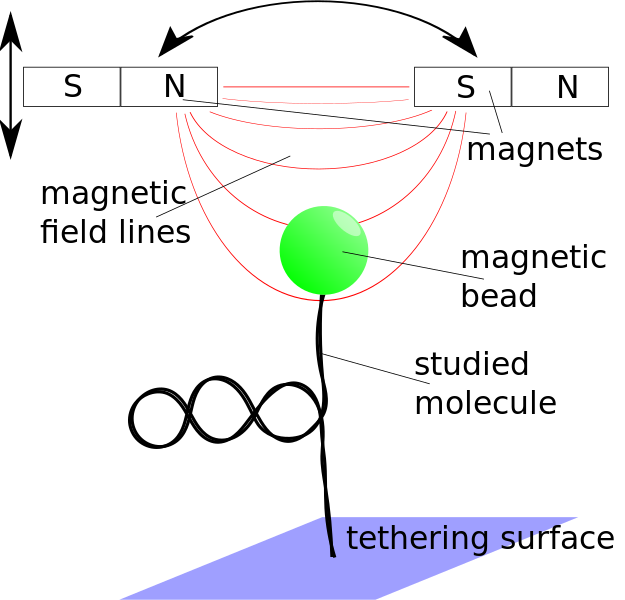
Steve Kenyon will be working on building a magnetic tweezers setup from scratch. What are magnetic
tweezers you may ask? In magnetic tweezers, one uses small (but strong!) permanent magnets (think refrigerator magnets on steroids) to pull on small magnetic beads that are attached to the object of interest. In our case we often are pulling on DNA or DNA with other proteins interacting with it. By carefully calibrating the machine one can measure exactly how long the molecule of interest is when you pull it with a certain force. By measuring how hard it is to pull the molecule straight, we learn something about the structure of that molecule before we started pulling on it. (Just like you can tell how tangled a telephone cord is by how hard it is to pull the cord straight.) (I know those of you who are younger out there may be wondering what a “telephone cord” is. Pleas ask your parents.)
This is a pretty involved project that will involve writing a lot of computer code in a funny computer language called Labview and doing a lot of interfacing of physical instruments, motors, and cameras with the computer and the program that is running it. All-in-all a great introduction into machine design!
So, I hope you will all enjoy our journey this summer with us and hopefully will be cheering us on as we race to get all of this done before classes begin!
The Summer Begins Anew
 What an exciting time! This summer we have one returning researcher, John Giannini, and two new researchers, Olayinka O. Fasawe (Fash) (below) and Ben Constable (right). These two will be taking the reins on the nucleosome work that I just got funding for through the Research Corporation Cottrell College Science awards. They will be looking at all of the interesting ways that ions interact with nucleosomes and how these ions sometimes manage to make the nucleosomes behave in peculiar ways. John will be continuing his research on DNA condensation.
What an exciting time! This summer we have one returning researcher, John Giannini, and two new researchers, Olayinka O. Fasawe (Fash) (below) and Ben Constable (right). These two will be taking the reins on the nucleosome work that I just got funding for through the Research Corporation Cottrell College Science awards. They will be looking at all of the interesting ways that ions interact with nucleosomes and how these ions sometimes manage to make the nucleosomes behave in peculiar ways. John will be continuing his research on DNA condensation.
In addition to helping them all with this work, I’ll be working on a project that uses x-ray scattering to study these same nucleosomes. It should be a busy summer!
But with this talent and the wonderful resources available to us, we’re ready for it!
Many general lab updates
For our nucleosome project, we have had some setbacks, most of which we have recovered from. As you remember from last time, our column had broken. That is still the case and we are still waiting for a replacement as it is on back-order. What I had failed to mention was that the sample itself had also had some issues. Namely, there was a white powder that had precipitated out of the sample and to the bottom of the tube. This was disturbing as I was relatively sure that this was all of my nucleosomes aggregating and falling to the bottom of my tube (something they are only supposed to do when I want them to). Since we were stuck without column, I thought I would investigate this issue.
It turns out that nearly all of my nucleosomes had aggregated. After some investigation, it turns out that the nucleosome sample was actually in 10mM Calcium Chloride (a +2 ion) instead of 1mM. This is an issue because nucleosomes aggregate at about 5mM of +2 ions. It is also a problem because the EDTA we add to stop the digestion is added to stop 1mM of CaCl2, not 10mM.
The first thing I needed to do was get the nucleosomes back in solution. We did this by dialyzing the nucleosome solution (and the precipitate) with 150mM NaCl (a +1 ion). This is done by putting the nucleosome solution in a bag that only lets ions and water pass through it, but not the nucleosomes. It turns out, if you add enough +1 ions, they will displace the +2 ions at which point the nucleosome should go back into solution. After a day of dialysis, we had significantly increased the amount of nucleosomes in our solution. After a weekend of dialysis, we had gotten back basically all of our nucleosomes. Now we had to make sure that they had been digested correctly (especially checking for over-digestion which shows up as smearing on our gel).
The results look like this. Now the furthest column to the right is our DNA ladder. This is used to let us know how long our DNA is. You match the known length of the ladder band to your sample. The bright band at the bottom is a 100 base pair DNA. Each step “up the ladder” is a 10 base pair step (110 base pair, 120 base pair, etc). The lane immediately to the left of the ladder is our sample. If you look and count carefully, you can see that it lies between the 140 base pair and 150 base pair “steps” of the ladder. This is perfect as the DNA should be 146 base pairs! Even better, there is no “smearing” to lower parts of the gel, which would indicate over digestion. You can see an example of this in a different sample that was run in the lane immediately to the left of our sample.
So as a wrap-up, we have the same amount of nucleosome we started with, all in solution and digested to the correct length. All that is left to do is to separate out any small amounts of double or triple or larger nucleosome arrays and we will be done. Since we are still waiting for our column to arrive, we may end up doing this through a sucrose gradient instead.
Not to be shown up, John is busy plowing through the ton of data that he has taken. Now that we have all of the data, we need to figure out why some of it looks great and some of it looks not-so-great. Things looked pretty bad at the beginning of this process, but each time I check in with him, he seems to have found another correction to make the data fall in line. If all goes well, we’ll have it all whipped into shape by the end of this week.
Great Successes and Amazing Failure
A lot has happened since I last posted. Let’s start with the positive.
We have successfully run a very nice non-denaturing poly-acrylamide gel. The key was to increase the amount of sucrose in or loading solutions. This helped minimize diffusion of the samples before they started running. Another thing that helped was a mistake I made. In my haste to make the gel, I accidentally inserted a 10-well comb rather than a 15-well comb. However, this actually resulted in much nicer results.
We see exactly what we would like. The first column is our normal DNA ladder. The second is the undigested chromatin. As you can see, it is all at the top of the gel. Then we start digesting. As we go to the right, the time of digestion is increasing. As you can see, by the sixth column or so, the size doesn’t change much for a few columns. This is the digestion being stopped right at the nucleosome core. After a bit of time, however, the digestion proceeds past this point in some of the nucleosomes, digesting the DNA that is around the core. That is why the gels get smeared towards the bottom for the last three columns. When we do our final digestion, we will make sure that we don’t let it digest this long.
After this great success, we digested one of our samples and were going to pass it through our size-exclusion chromatography column. This column will separate our sample by size. This will allow us to collect all of the nucleosomes that are the exactly correct size, while ignoring those too big or anything that is too small (bits of DNA, etc.). The setup is shown to the right.
One thing about a size-exclusion column is that it must never dry out. The tricky thing is that you need it to almost dry out to add your sample so that your sample stays relatively concentrated in the gel. To make a long story short, I brought the column to the almost dry level and the closed the valve. However, the valve leaked slightly and after 15 minutes, when we loaded the sample, I noticed the gel had completely dried out.
At this point we needed to recover the sample (which we did right away). We then took the column back to the lab to recover the media (the white stuff in the column) so we could rehydrate and repack the column. During this process, I overtightened the column in the vise that was holding it. So now we are waiting on a replacement column and replacement media. It’s funny how much a small leaky valve and 15 minutes can change things.
And more DNA gels
Well, we keep getting better at least. This digestion showed a much better digestion. As you can see, the 0 minute digestion showed a smear of DNA, while all lanes that were digested for 5 minutes or more showed a nice single band that can be believed to be around 146bp. As can be seen we have a few issues.
First, the digestion is supposed to take around 30 minutes. This is helpful because it means that you can make sure that you are just digesting to 146bp and not overdigesting. Ours is taking 5 minutes. In our next digestion, we’ll try decreasing the amount of nuclease (the thing that “eats” the DNA) by a factor of 10. Hopefully we’ll then see a little more of a progression and less of a step function.
The second issue is that the gel still doesn’t look great. The first issue is the ladder. We obviously didn’t run long enough: Our 100bp standard only went about 1/2-way down the gel. However, conditions were controlled enough the last time that we should be able to have a nearly-perfectly timed run.
The other issue with the gel is that the DNA in all but the ladder lane is quite diffuse. It would be much easier to read if they looked like lines, similar to the ladder in previous gels. My theory is that this is due to the sample not being dense enough when loaded and therefore diffusing before it starts running in the gel. An easy fix for this will be to increase the amount of sucrose before loading these samples.
Finally, a problem with the gels that I’m not showing you is that I messed up the loading of the other gel. This is actually Travis’s gel. Mine didn’t have a ladder! Well, I’ll have to do better next time.
Things with John’s project are going quite well as you can see by his posts. We should have nitrogen tomorrow which will allow him to run more samples. After that starts up, we should be on our way to having all of the DNA aggregation samples done by the end of next week if everything goes well.
New Gels, New Challenges
Note: I realize these posts may be confusing to those outside of biology/biochemistry fields. In general, gel electrophoresis separates molecules by size: In this case, biggest are at the top and smallest are at the bottom. For more, look at this great animated introduction.
As you remember from my last post, our last gel ran quite well, but the results showed that we had failed to digest our inter-nucleosome DNA. This time, we modified our digestion process (and by modify, I mean went back to the process I had been using before I decided to “improve” my process).
 This time however, we have another issue. The gel itself gave us problems. There were a couple issues layered on top of one another, but the main one was that our power supply that runs the gel decided to quit some time in the middle of the night. This had two effects, one was that the DNA had time to diffuse and migrate in any old direction it wanted to during the time that the power supply was off. This causes a broadening of the bands which I think we are seeing here. Another issue was that it forced us, due to time considerations, to ramp up the voltage to very high values which I think is what caused the intense upside-down V-like shapes to the bottom of our gels. Finally, we ran the gel a little too long losing our 100bp marker. In the previous post, you could see the 100bp and 330bp markers, but I am pretty sure that the marker you see in the first lane is simply the 330bp marker. This makes it impossible to determine as, even though some of the middle markers are resolved, we have no way of counting up from the markers (each sub marker is 10bp).
This time however, we have another issue. The gel itself gave us problems. There were a couple issues layered on top of one another, but the main one was that our power supply that runs the gel decided to quit some time in the middle of the night. This had two effects, one was that the DNA had time to diffuse and migrate in any old direction it wanted to during the time that the power supply was off. This causes a broadening of the bands which I think we are seeing here. Another issue was that it forced us, due to time considerations, to ramp up the voltage to very high values which I think is what caused the intense upside-down V-like shapes to the bottom of our gels. Finally, we ran the gel a little too long losing our 100bp marker. In the previous post, you could see the 100bp and 330bp markers, but I am pretty sure that the marker you see in the first lane is simply the 330bp marker. This makes it impossible to determine as, even though some of the middle markers are resolved, we have no way of counting up from the markers (each sub marker is 10bp).
As for the digestion itself, things look pretty good. We got rid of our smear of DNA and it is obvious that the DNA is migrating to approximately the correct place. There are only two issues I am worried about. One is that aside from the zero time point (the second lane), most of the lanes look the same. In other words, it seems as if we are completely digested by 5 minutes.
The second issue is that the lanes that do look different are completely random. The corresponding times are 20, 40, and 50 minutes. By 50 minutes, we shouldn’t see anything above that band of brightness on the bottom. The fact that it is random seems to indicate a possible pipetting error. However, I ran and loaded two separate gels, so the pipetting error would have had to have come earlier. We’ll try going over our process to make sure that this isn’t an issue.
Other than that, it’s back to do the exact same thing again. This time we’ll be using a different power supply so we don’t run into the same problems.
DNA Gels and Poor Digestion
As Travis mentioned in an earlier post, today we finished running our gel. With the help of the Steve James lab, we imaged our DNA. The results were not promising.
What you see on the left is just the DNA ladder. Those numbers correspond to the number of base pairs (bp). On the right, in the lane labeled 1, you see our DNA ladder with red lines tracing the corresponding markers. As you can see, we can clearly see the 330bp and 100bp markers. What we can’t see are all of those nice little lines in between. This could be a problem in the future, but is small beans compared to the mess further to the right.
In lanes 2-12, we had placed DNA samples that had gone through various times of digestion. What we were trying to do was take DNA that was all kinds of different lengths and snip most of it down to 146bp. Each lane corresponds to 5 minutes later than the one before it. So lane 2 is time zero, lane 3 is 5 minutes of digestion, lane 4 is 10 minutes of digestion, and so on. Now, you don’t have to be an expert to realize that lane 2 looks identical to lane 12! This is a bad sign. But what does it mean?
There could be many issues: The gel could have been run incorrectly, the samples could be contaminated, or we could have so much DNA (gels aren’t normally this bright) that it somehow makes us unable to see what is actually happening. But I’m pretty sure none of these are the cause.
The procedure for digestion I used was a modification to the procedure I had used the first time I did a nucleosome preparation. The modification required a few more measurements, but it was claimed in the paper I pulled it from that this procedure gave better results (i.e. more nucleosomes at the end). We did these extra measurements (I believe correctly) and added the amount of nuclease (the digestors or eaters of the DNA) that the new procedure called for. However, as both Travis and I found out in later calculations (that perhaps should have been done before the trial digestion) this new procedure called for 100 times less nuclease than the former procedure called for. The final result: No digestion.
Luckily enough a) We were doing a trial digestion, not the real thing, so we didn’t lose any real amount of samples and b) It is quite easy to repeat the trial digestion with the old method and then rerun our gel. And that’s exactly what we are going to do tomorrow.
Wish us luck.
Ordering and thinking
I’ll let everyone reading this blog in on a little secret. A good portion of a researcher’s time is spent on menial tasks and tasks that look very similar to “doing nothing”. Today was one such day for me (as was much of the week). I spent most of my time ordering supplies, running around borrowing/stealing supplies from others, and planning our future projects. This last one was the most difficult, as my brain has just not been cooperating lately. People will say that this is related to the 1-month-old baby in my house, but I personally just think it’s just the consequence of a more-hectic-than-normal week. To at least a little justify my presence in the lab, I poured a very dangerous powder, PMSF, into isopropyl alcohol to make a slightly less dangerous solution. (Less dangerous because it’s a lot harder to inhale a liquid than a dust.)
In real news, we are going to be doing two rather important things next week. First, we are making our nucleosomes. This is exciting, although perhaps a bit early. It’s early because it is questionable whether we will have everything we need ready by that time. However, in science it is often the case that you must go ahead with plans and hope for the best. If you wait until everything seems to be ready, you never get anything done. The good news is that our Fisher rep claims our UV detector is coming on Monday. With some luck and some help from Prof. Brandauer in Health Sciences, we should be able to get that up and running on Monday. Additionally, I’m hoping to make all of the stock solutions we need on Monday so that we aren’t running around trying to make those all last minute. Our collaborator, Xiangyun Qiu, is coming on Tuesday to see the prep and generally just to visit, so it this coming week should be busy.
The other exciting thing is that I’m expecting to start running the condensed DNA samples that John has been preparing the machine for. Although he’s still having a hard time nailing down that last 2-3% of systematic error, I think we’re pretty close to being at a place that we can run at. I’m guessing that come Friday or so, we’ll be in business. As Xiangyun is the one that gave us these samples, we’ll also take some time during his visit to talk to him about the samples and our general running procedure.
So it should be an exciting first week of “summer”.
We’ve got blood!
At 10:00am this morning, Travis and I arrived at the Berry Blossom Farm. We went into their unassuming store front, the entire farm complex smelling of chicken (and I don’t mean the fried kind). In the store, I asked for Alden and told them that we were hear to collect chicken blood. She seemed to have been informed of this possibility and went back to get Alden.
Alden came out dressed in quite messy clothes and a rubber apron. He said that we should come on in and get our blood. I informed him that our equipment (basically a styrofoam container filled with ice, 2 beakers, 50ml of 6% sodium citrate, and cheesecloth) was back in the car. As I walked back, I took a cue from Alden’s appearance and removed my outer shirt so I was just wearing my undershirt. This was to be one of the smarter things I did today.
Alden escorted us through the maze-like complex until we got to a small room. The first thing I noticed about the room was the intense smell. It had an intense animal smell (imagine a room with 50 wet dogs) combined with undertones of blood. The scene seemed to be complete chaos with birds in various states of life and death (alive, dying, dead, plucked). A man who was doing the slaughtering moved quickly removing the dead chickens, putting in new ones, and rapidly dispatching these new be-coned birds with a swift twist of the knife in their throats.
A steel fixture with four steel cones each containing a dying/dead bird. Under the cones was a trough that caught the blood. To the left was the plucker. Basically, this consists of a washer machine type contraption, only there are numerous plastic “fingers” that stick out and remove the feathers as the chicken tumbles around inside. Due to the presence of this machine, feathers were continuously flying throughout the room.
Once we got inside this room, the chaos was infectious. Alden used a garden hose to wash down the cone we were going to use. I quickly asked Travis to pass me the largest beaker and the sodium citrate (which prevents clotting). I noticed a naked chicken come flying out of the plucker and land on the floor, I don’t know if this is by design or if it simply escaped the plucker’s grasp. I dumped the sodium citrate in the beaker. The next chicken was loaded; I put the beaker underneath the chicken and with a quick stab and a twist of the butcher’s knife, the blood started flowing. I collected the slow drizzle of chicken blood. It took three chickens and about two minutes to collect 200ml of chicken blood, even with the butcher turning the head of the chicken so I could get every last drop. One minute through the collection, the women in the next room in charge of the last bit of cleaning of the chickens started singing. (Travis thought it sounded like chanting.) The clear tones of the women’s voices clashed strongly with sight of chicken heads and blood on the floor.
Travis and I exited the building. We poured the chicken blood through a cheesecloth to remove the feathers and other various non-blood elements and put the cleaned blood on ice. As we were leaving to go back to Gettysburg to extract the red blood cells from the whole blood, we were stopped by Alden.
“Do you have a way to keep a chicken until you get back?” he asked. “We have a few extra.”
“Sure,” I said. So Travis and I returned to Gettysburg having bringing back some chicken blood, a frozen chicken, and experience we probably won’t forget for a while.





