Welcome back to the Andresearch blog! This week hasn’t been very exciting, so sorry in advance if it seems repetitive, but when you’re doing research, all you do is repeat your work until it is good. The gist of the week was washing DNA samples to do more runs in the ITC.

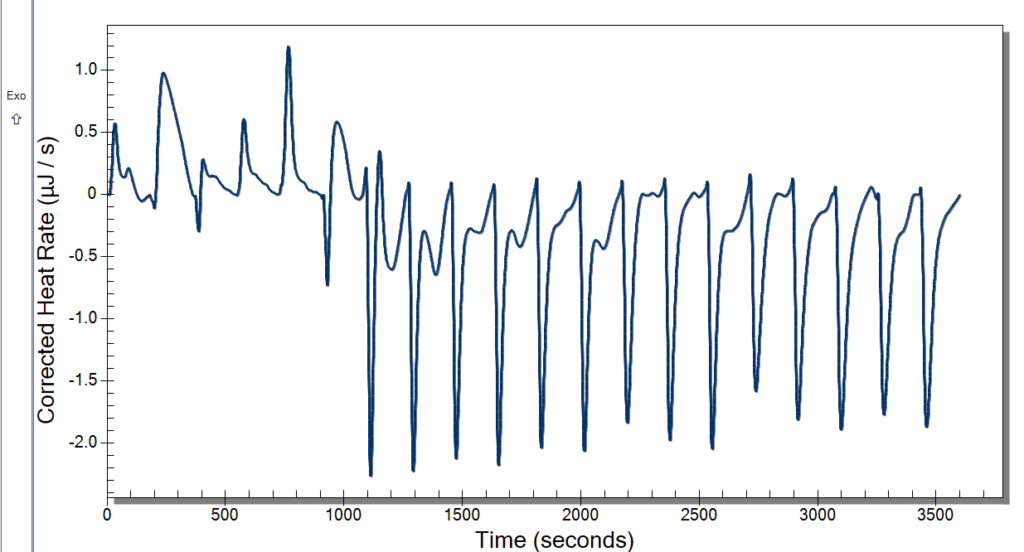
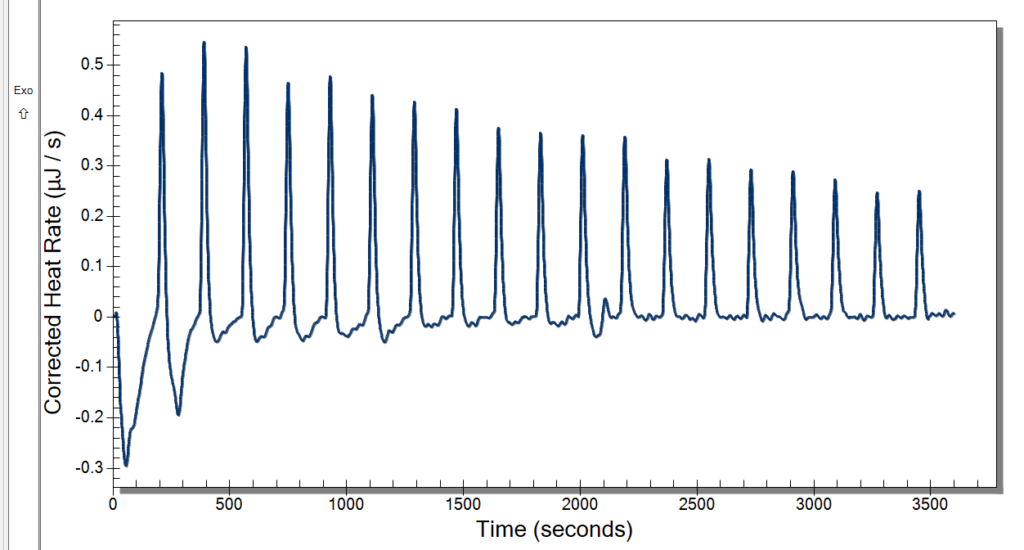
Wednesday, we learned new procedures regarding using the ITC. We now use a detergent called hellmanex to clean the cell because our samples are quick sticky and need a little more than water to get it out. We thought we had some really good data after changing the settings previously mentioned, and the extensive cleaning might have also helped. Our data looked pretty nice to us, but once checking the analysis software, it wasn’t what we were looking for. We discussed potential issues, and we decided to up our concentration of CoHex to 30mM.

We plan to make DNA gels in order to find the length of our DNA, so we prepped for that by making 4 samples with varying dilutions. We had one at our normal concentration, and then a 10x, 100x, and 1000x dilutions. We made two things called ladders as well which you use to gauge the lengths after making the DNA gels.
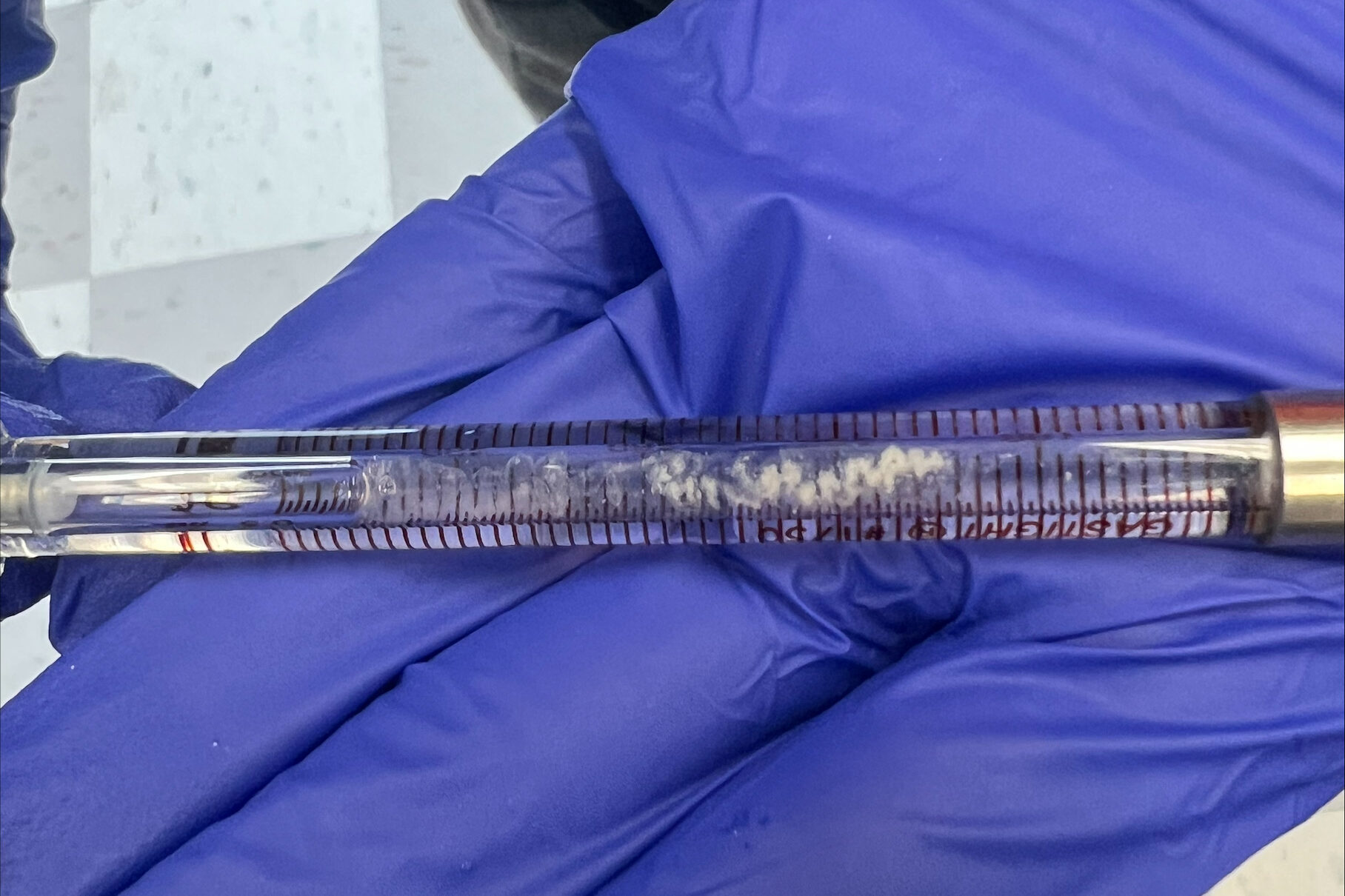
When washing more DNA on Monday, we used a new type of filter, and our data was kind of weird. These new filters didn’t give us as much DNA than before, and we had to add more NaCl. We kept these in a separate tube to see if this poses an issue or not. This washing used up the rest of Batch One DNA, so on Tuesday we made Batch 2 after doing our ITC runs. Our ITC run seemed to give better results, but we were left with the DNA all clumped up in the cell as pictured below. We weren’t able to ask if this was what we were looking for or not, so we only did one run. For Batch 2, we followed normal DNA making procedure.