I started out this week by trying to purify the DNA-NP conjugates that I made last Friday. Unfortunately, all of the samples except the 1 microgram/mL DNA sample showed significant aggregation after sitting at room temperature for the weekend. I chose to only attempt purification of that one sample but after just 2 spins in the centrifuge the sample had completely aggregated out. I tried the same concentration with slower spin speeds and longer times but the sample continued to aggregate out. Thinking that perhaps the conjugates were forming histone-like aggregates, I next tried digesting the “nucleosomes” with the micrococcal nuclease, an enzyme Sarah is using to isolate mononucleosomes, and then purify them. The result was a low concentration solution with lots of black flecks of aggregation that floated in solution and resisted pelleting following extensive centrifugation.
 |
| If you look closely you can see the black flecks of aggregation |
Unfortunately in the Thompson lab, they ran out of nanoparticles and the CTAB needed to make new nanoparticles so instead I turned my focus to shearing down the calf thymus DNA using sonication so it would be less likely to incite aggregation. I had attempted this in the spring but had not had any success mostly due to the fact that I had to use a water sonicator that is not optimised for DNA shearing. The protocol called for a probe sonicator. There is only probe sonicator at Gettysburg in the Biochemistry lab and I had been told it was broken. However, with nothing else to do, we decided to take the probe sonicator and try to fix it. Turns out the sonicator was not broken, it worked just fine once we plugged it in. So I ran the shearing protocol I found in a paper I read. The sonicator is quite loud. I only ran it at level 3, but higher levels probably require ear protection.
After I ran the shearing protocol, I characterised the sheared and unsheared DNA using DLS, UV-vis, and gel electrophoresis. The gel electrophoresis showed that the DNA was sheared down from a 10 kb chain into a slew of differently sized fragments ranging from 1 kb down to much less than 0.5 kb. This data indicates that the shearing protocol was very successful in producing fragments small enough to hopefully resist aggregation.
 |
| DNA Ladder |
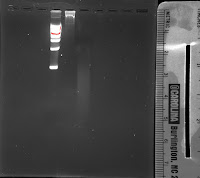 |
| Gel electrophoresis. The first lane is the DNA ladder, the second lane is the unsheared DNA and the last lane is the sheared DNA. |
Since I learned that new CTAB cannot be obtained until the middle of July, I am considering using negatively charged citrate gold nanoparticles and wrapping them in positively charged lysine molecules before mixing them with the newly sheared DNA.
 |
| Don’t do drugs kids. Except caffeine… lots of caffeine. |





















