Hey! I’m Matt, one of the researchers working with Dr. Andresen this summer. My project is a continuation of a published work Dr. Andresen collaborated on back in 2013 (link). Allow me to give you some background before I explain exactly what it is I’m doing. The electrostatics of nucleic acids are fundamental to both nucleic acid structure and function. The physical origin of DNA condensation -a process essential for gene regulation- remains unsettled. It is the goal of our research to further our understanding of both these aspects/processes by observing what happens to DNA condensation when the solution containing the DNA has different amounts of differently charged ions. The research Dr. Andresen worked on in 2013 explored what happens when the amount of +2 and +3 ions varies in the solution, my research is interested in +1 and +3 ions. Basically, we look at how many of each ion bind to the DNA as their concentrations vary. My job, more specifically, is to simulate the DNA ion-competition according to how the Nonlinear Poisson-Boltzmann (NLPB) equations say it should work. Practically, this means arranging the DNA strands into predetermined shapes (in our case a hexagon) to try and mimic a small strand of DNA in vitro. Hopefully, our research will provide more insight into, not only into DNA condensation, but also biomolecular electrostatics in general.
As for my work these last few weeks, it’s mostly been trying to learn (and then unlearn) a few python packages for working with .pdb files. .pdb files are files with specific formatting that stores information about the DNA like its positional data, total charge, connections between nucleic acids, etc. This started off pretty well with me making rapid progress in terms of positioning the DNA strands where they needed to be (see below), but sadly, as I learned this Thursday, the packages I was using to do this unfortunately break the .pdb files when they edit them. Not break them completely, mind you, but just enough where the accuracy of the simulation takes a serious hit and thus I had to abandon them. This left me with only one option: write my own. Fortunately, I don’t need much of the functionality of those packages, I really just need the ability to modify the positions of the strands. I imagine I’ll have this done sometime early next week as the only snag left is that .pdb files require a very specific format that is whitespace dependent and I’m having trouble getting everything to literally lineup as it should.
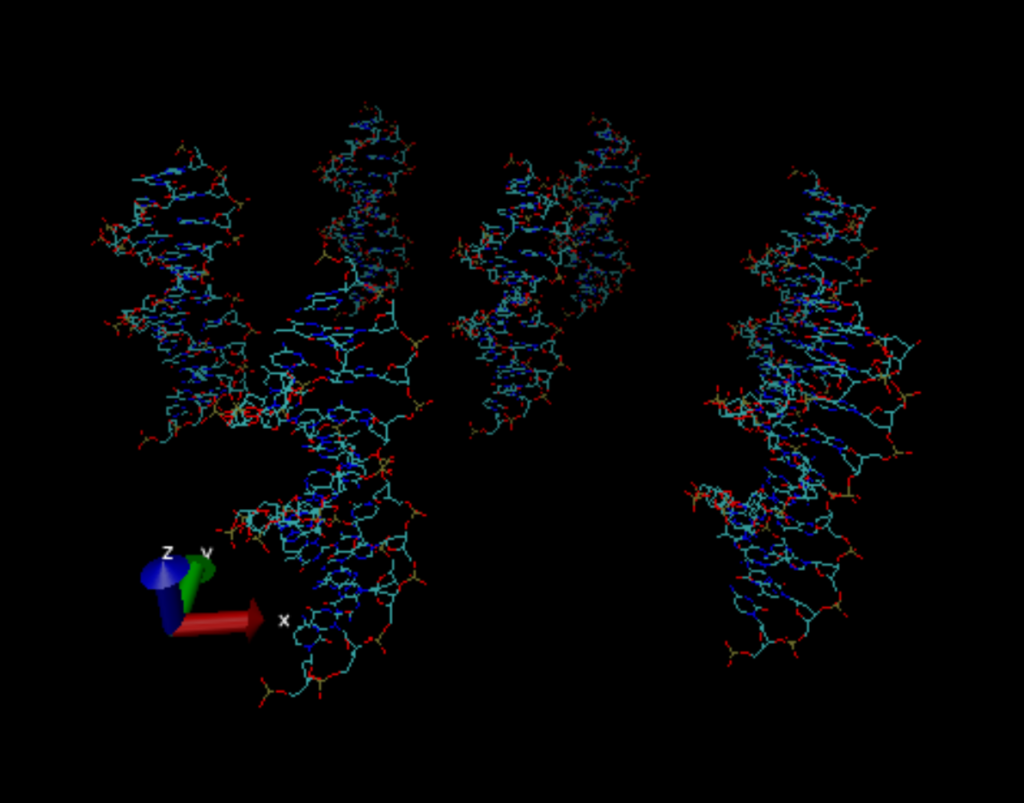
After I work out the kinks, the next step will be to run my .pdb file through https://server.poissonboltzmann.org, a free Poisson-Boltzmann equation solver. This allows me to outsource the processing of the data to a webserver so my poor laptop doesn’t overheat and die trying to do all that math. Once I get the data back and in a state that I deem acceptable, I’ll be able to start doing some actual analysis, but at this point that’s getting ahead of myself.



